technology
Request your free digital copy of Genome Engineering with Programmable Nucleases for Animal Model & Cell Model Generation
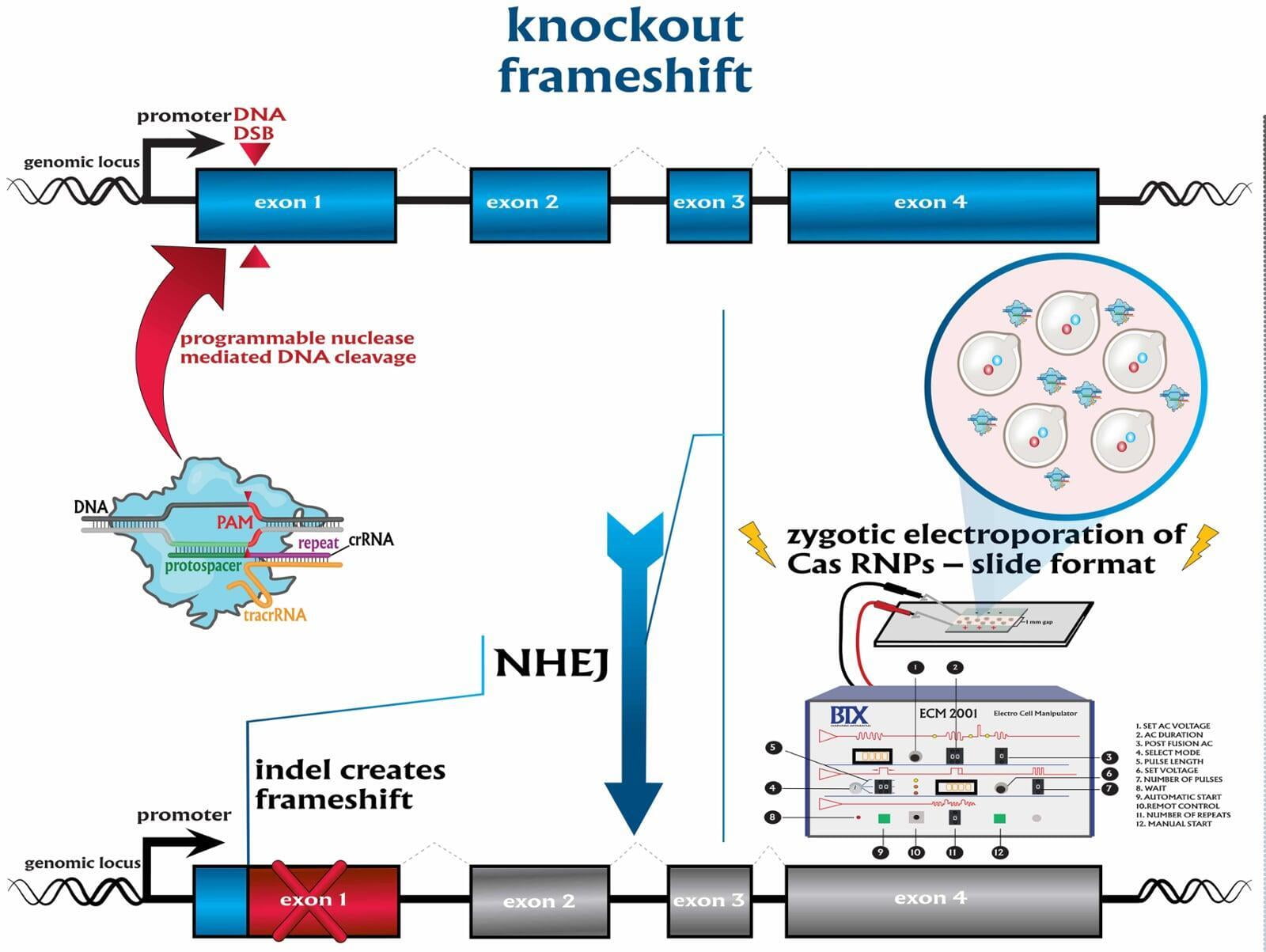
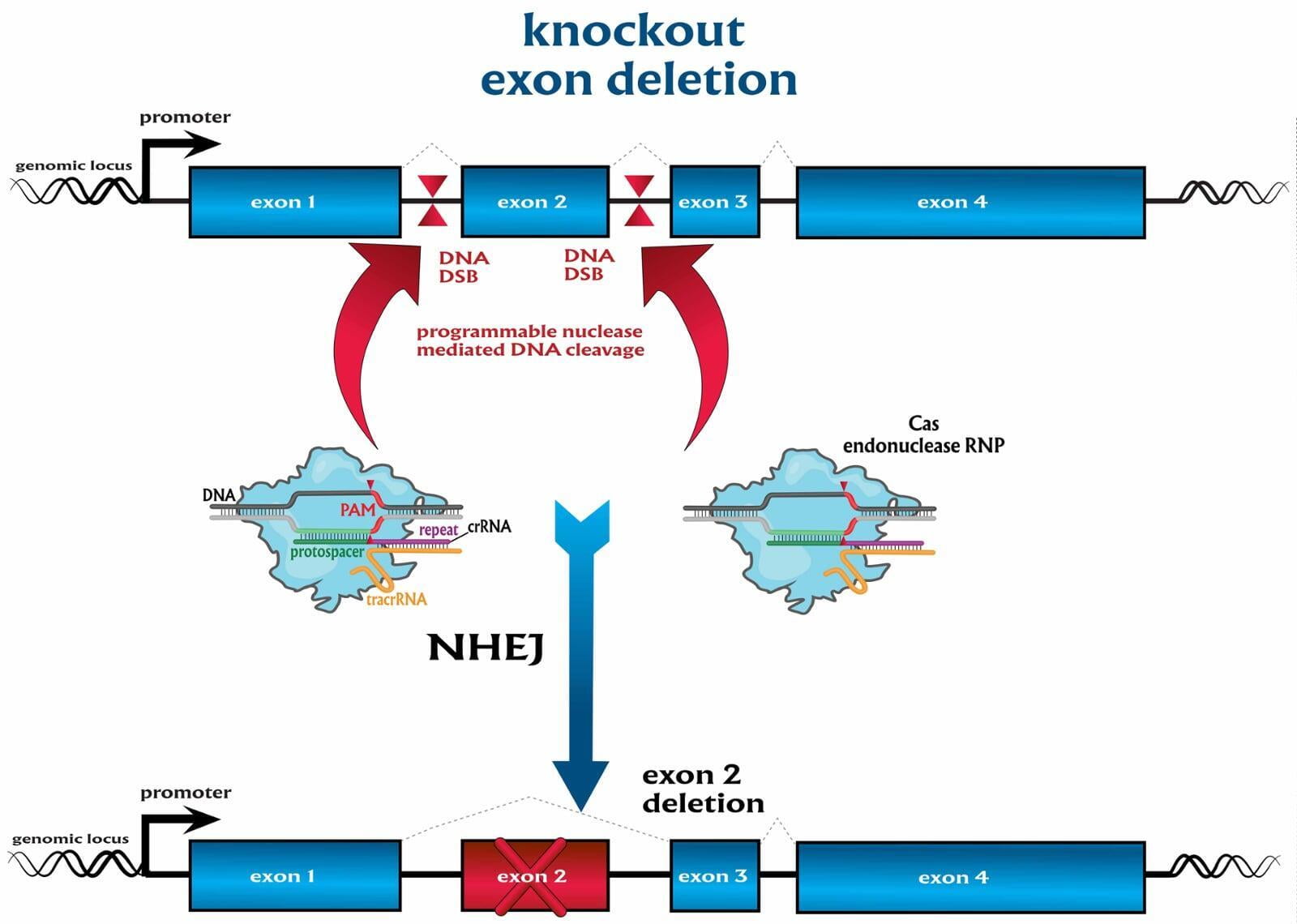
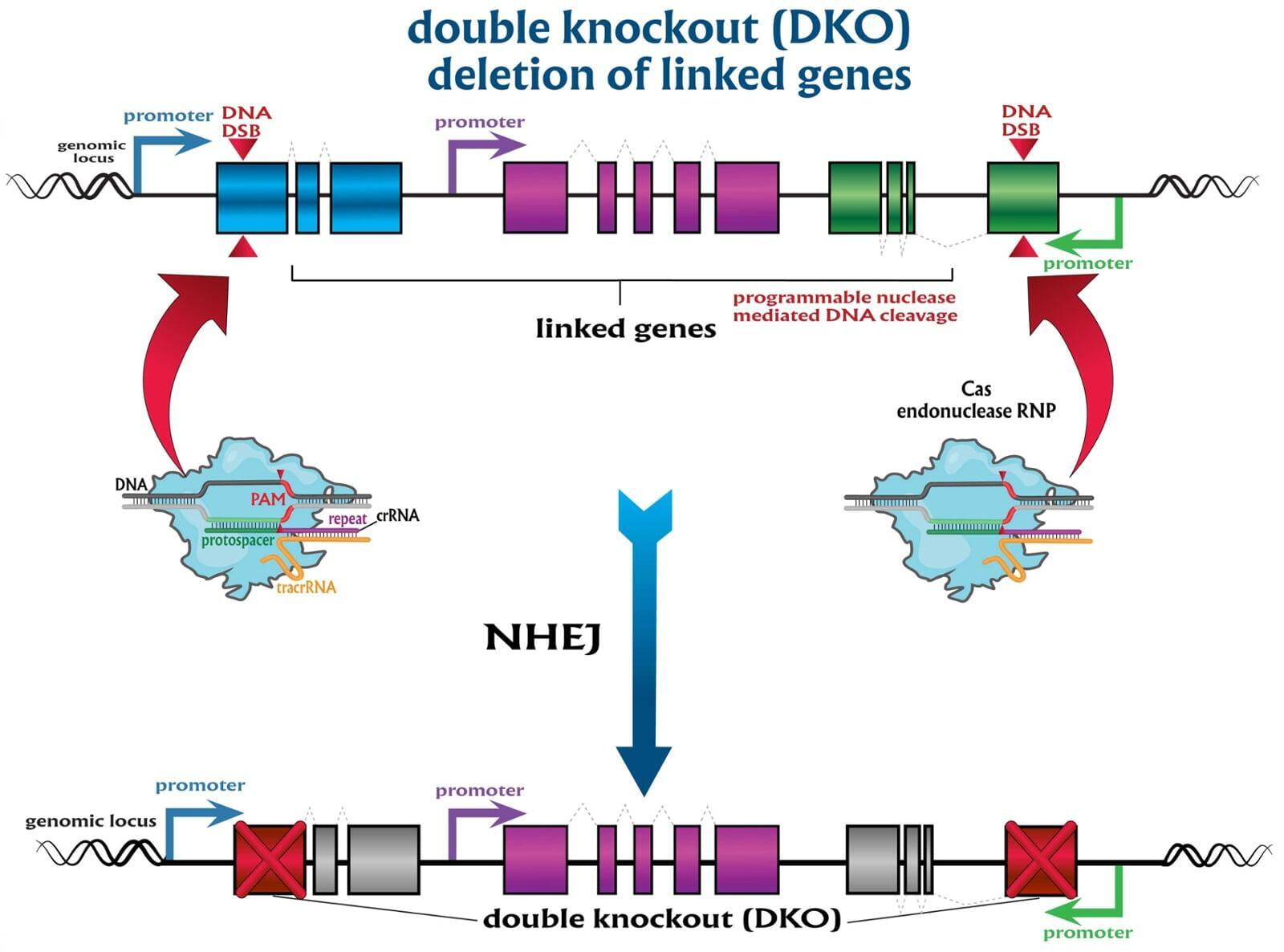
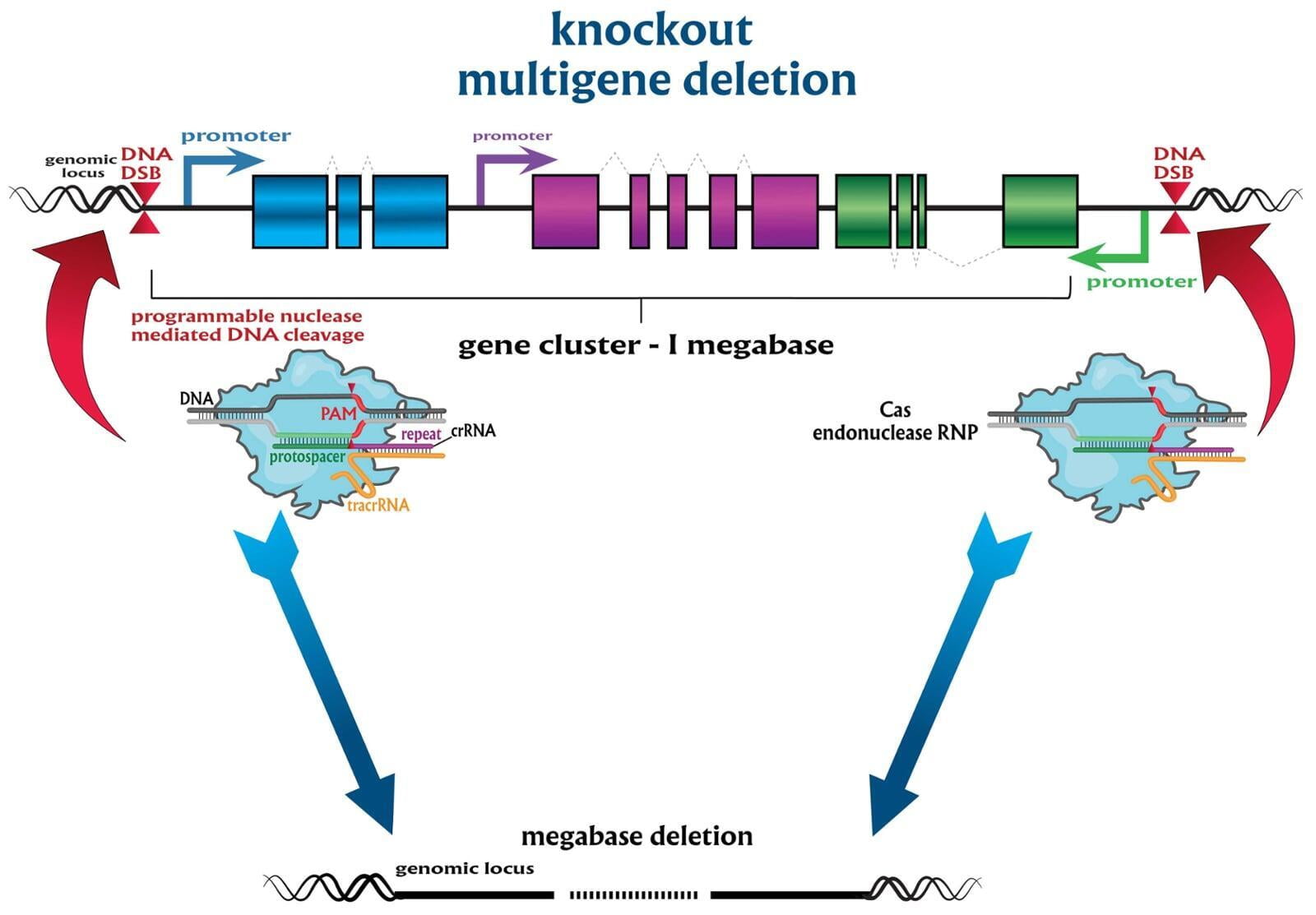
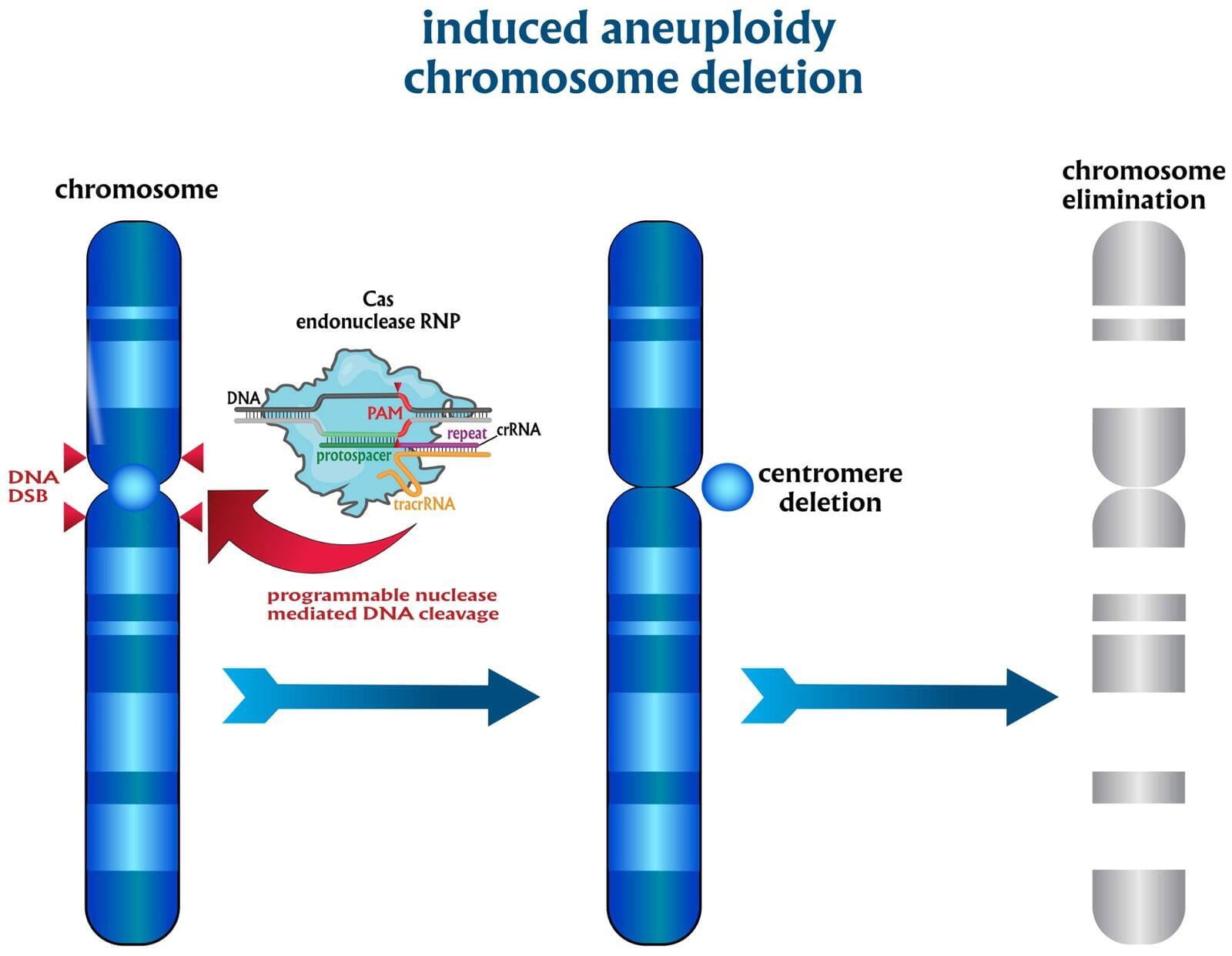
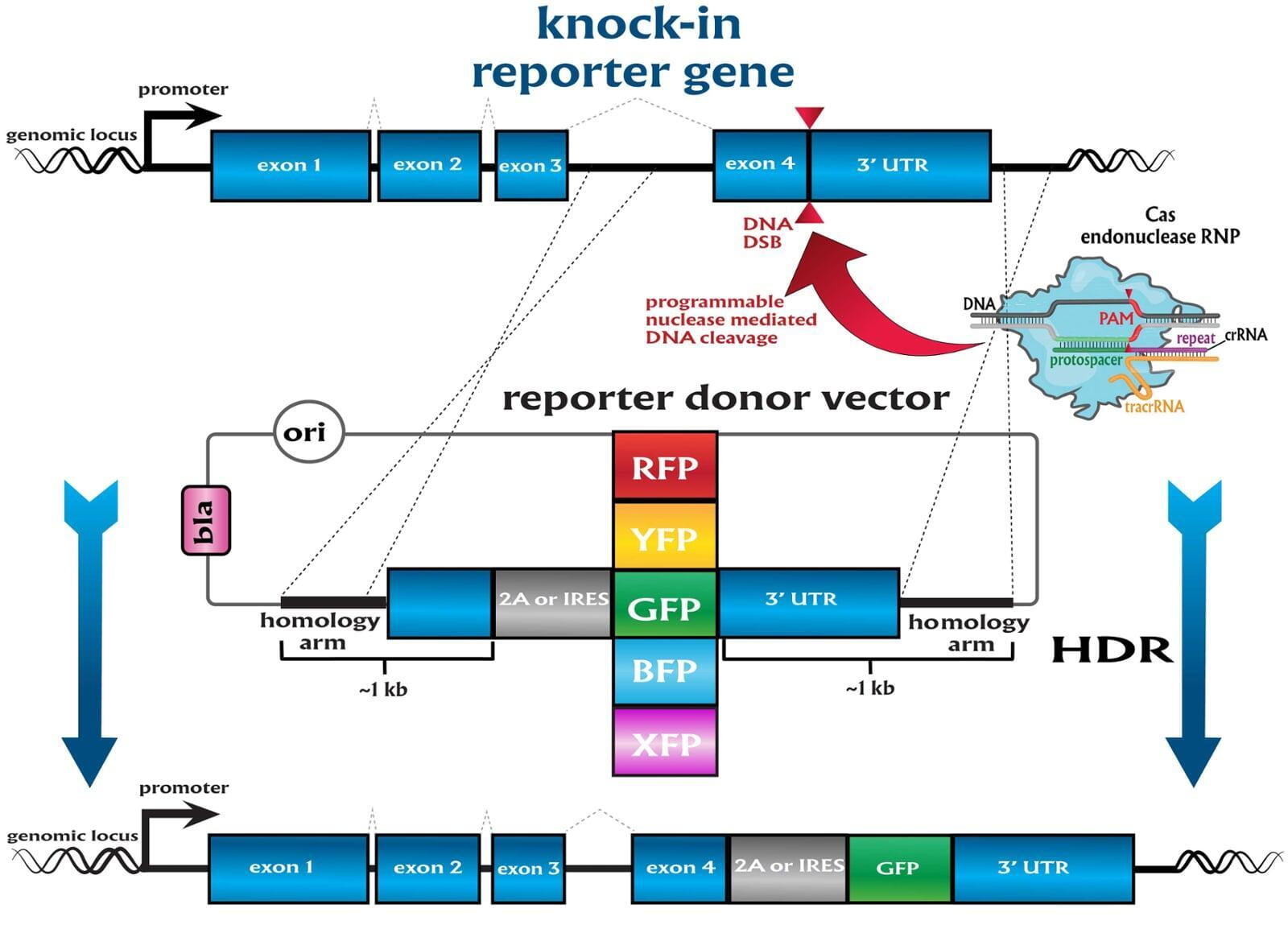
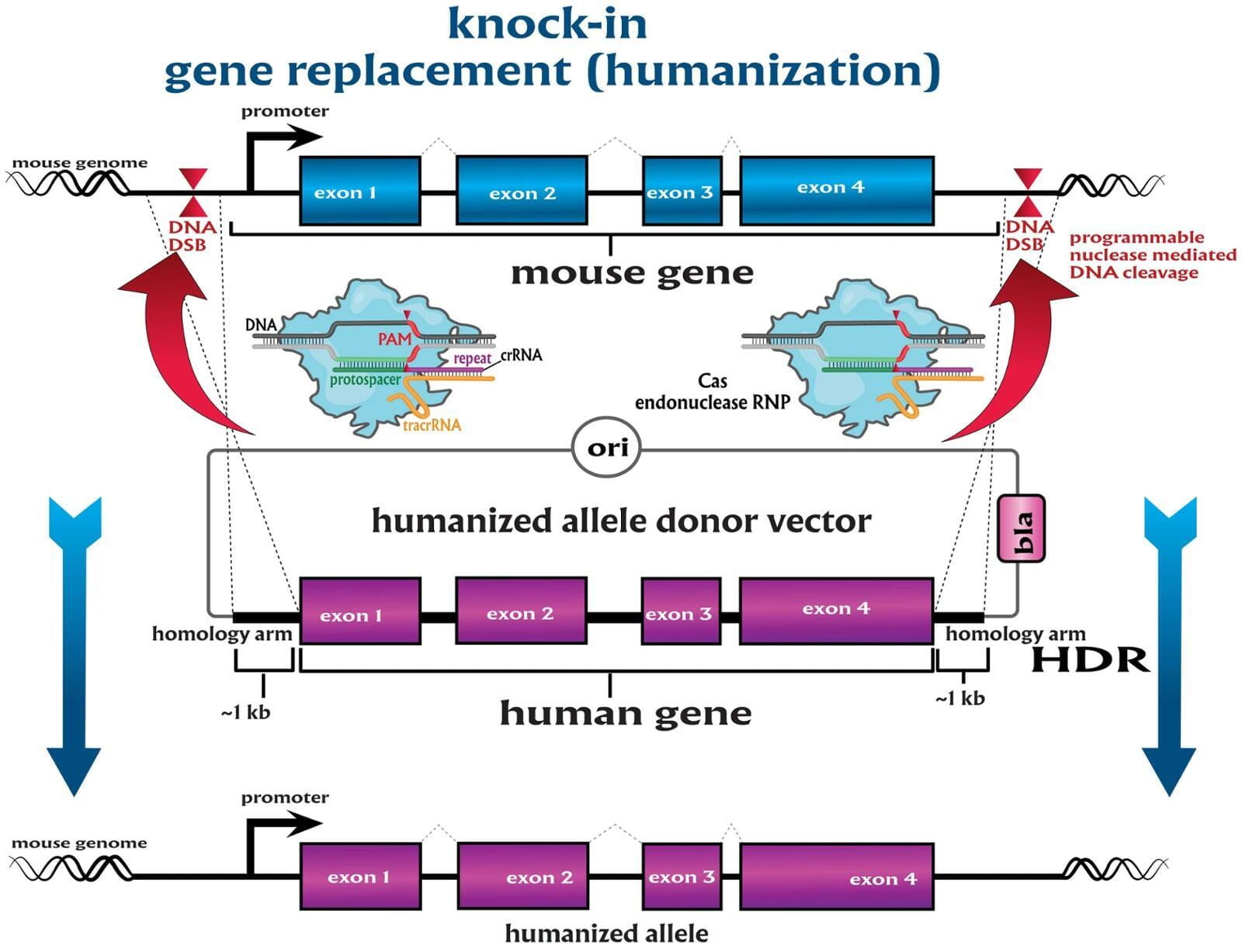
To advance the study of gene function, NovoHelix offers a comprehensive loss-of-function gene platform to generate the desired animal model. CRISPR-nuclease assisted gene inactivation services include disruption of the translational reading frame such as frameshift stops (indels) to create simple knockouts, exonic deletions, single or multi-gene deletions, double knockouts (DKO), deletion of linked genes, megabase deletions and whole chromosomal ablation (induced aneuploidy). For knockout of multiple genes simultaneously also known as multiplexed genome engineering, the Cas12a (Cpf1) nuclease processes a single CRISPR array containing several target sites and disrupts the reading frames of multiple genes (Zetsche et al 2017, Campa et al 2019¹,²). While this multiplexing technology is still in preliminary stages, NovoHelix has demonstrated expertise using cutting edge research tools to inactivate multiple genes and has entered the digital genome engineering era.
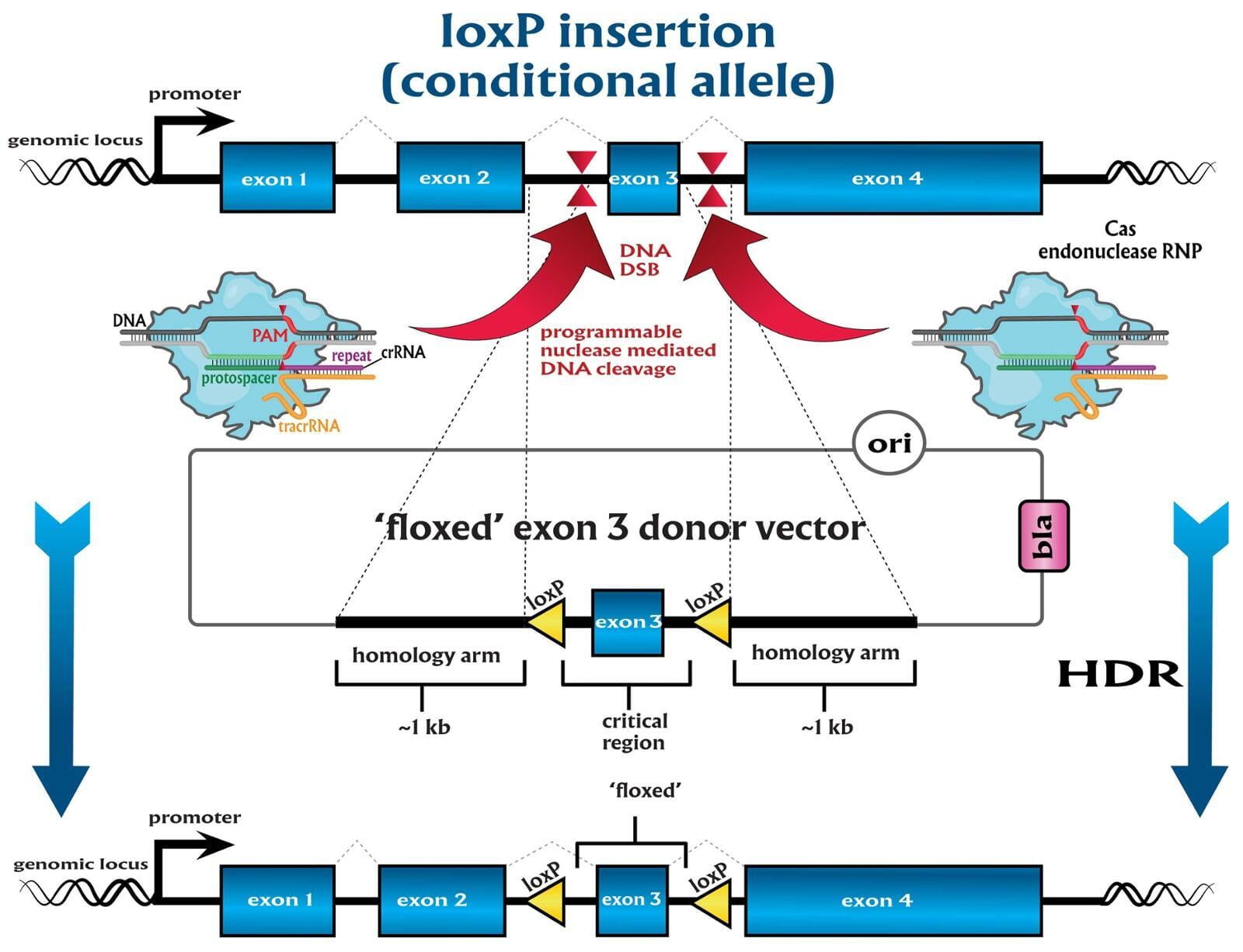
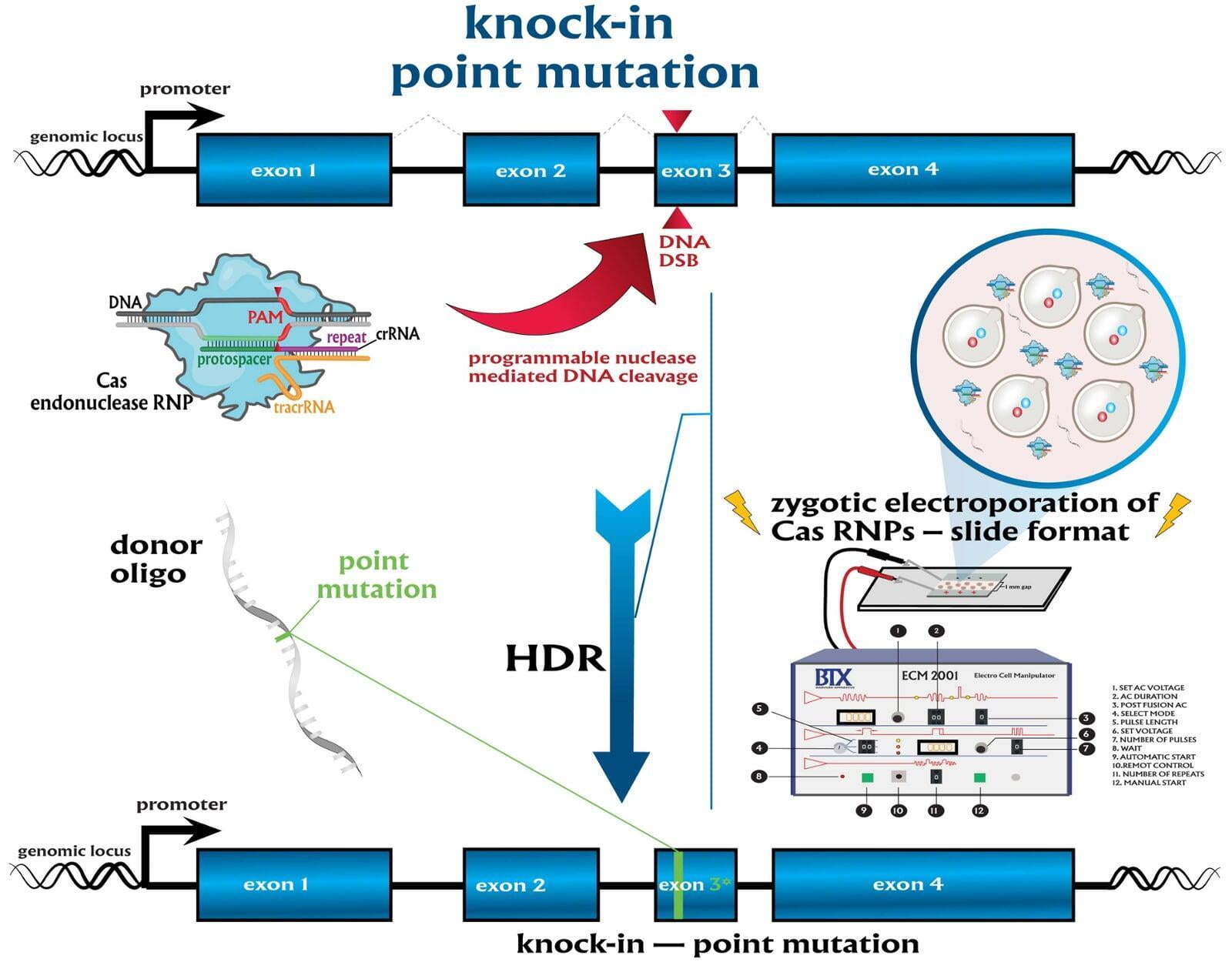
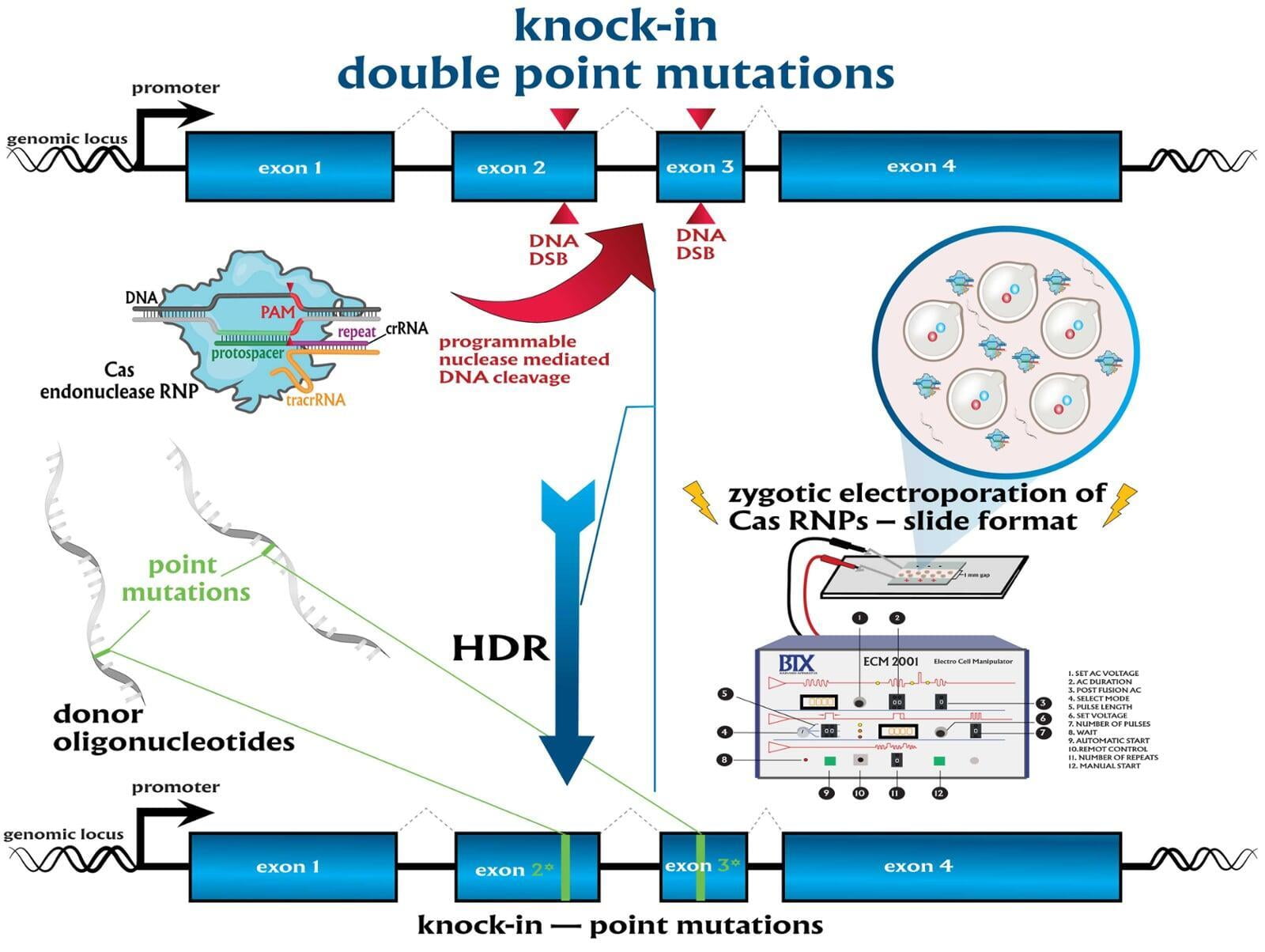
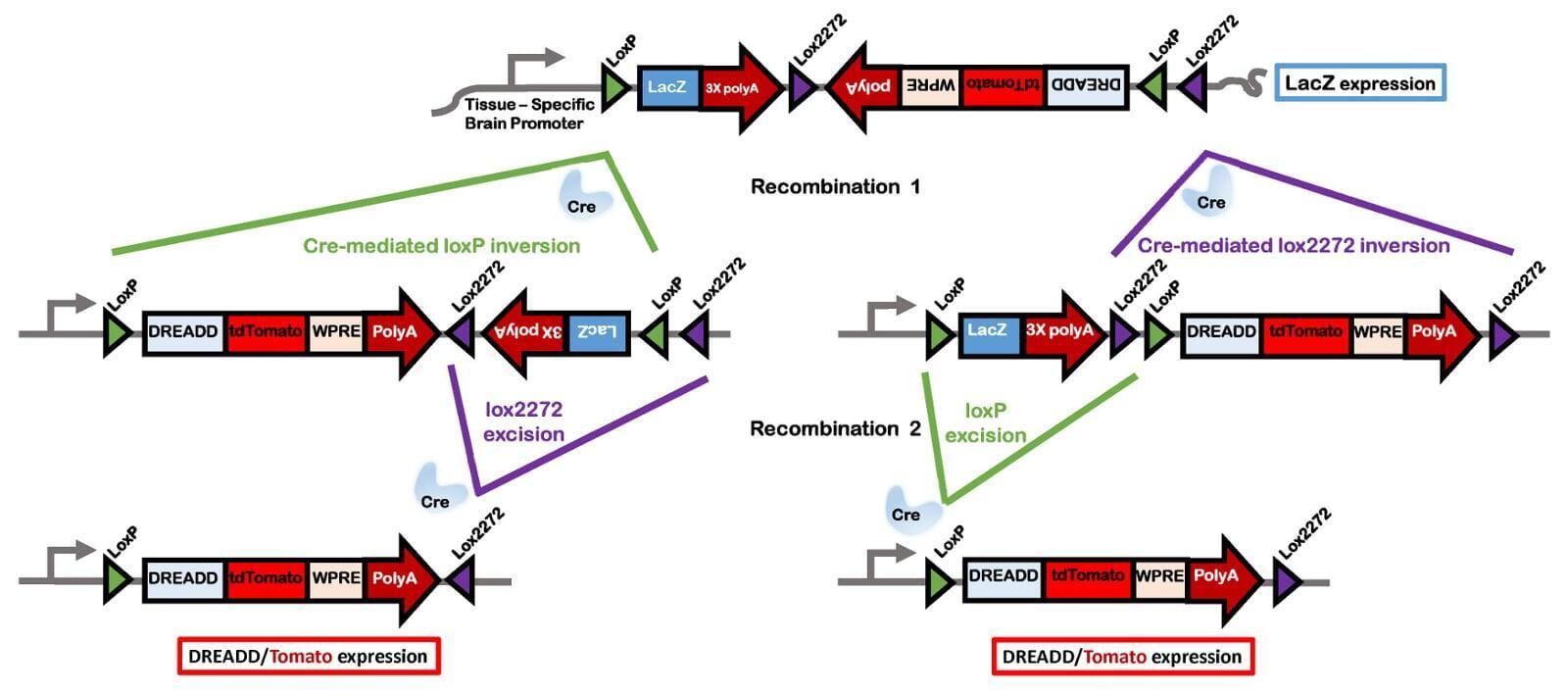
In comparison to direct knockouts, conditional knockouts (cKO) allow for temporal and spatial control of gene disruption. To generate a cKO, the target gene’s critical exons are often flanked with directional loxP sites termed ‘floxed’ which can be excised by Cre recombinase. Crossing a Cre-expression driver mouse to this floxed allele affords the investigator to conditionally excise the targeted gene only in cells which express the recombinase Cre. Conditional gene disruption is useful for modeling embryonic lethal genes, tissue-specific gene deletion or developmental-stage gene disruption. Our service packages include options such as guaranteed founders or pricing per microinjection session, flexible repair templates such as conditional donor vectors or long single-stranded DNA (lssDNA), and the choice of RNA molecule formats for guide RNA (gRNA) evaluation using the latest design guidelines: full-length guides, synthetic single guide RNAs (sgRNAs), chemically-modified crRNA + tracrRNA or sgRNAs, truncated sgRNAs, & hairpin-sgRNAs (hp-sgRNAs). The advantage of testing multiple high-specificity Cas endonuclease variants and RNA molecule formats allows NovoHelix to tune the activity of custom RNP complexes to produce the highest on-target activity in our surrogate cell-based gene editing assays while limiting potential off-target activity.
- conditional by inversion (COIN) as described by Economides et al, 2013³
- flip-excision switches (FLEx) as described by Schnütgen et al, 2013⁴
- invertible intronic cassette (FLIP) as described by Andersson-Rolf et al, 2017⁵
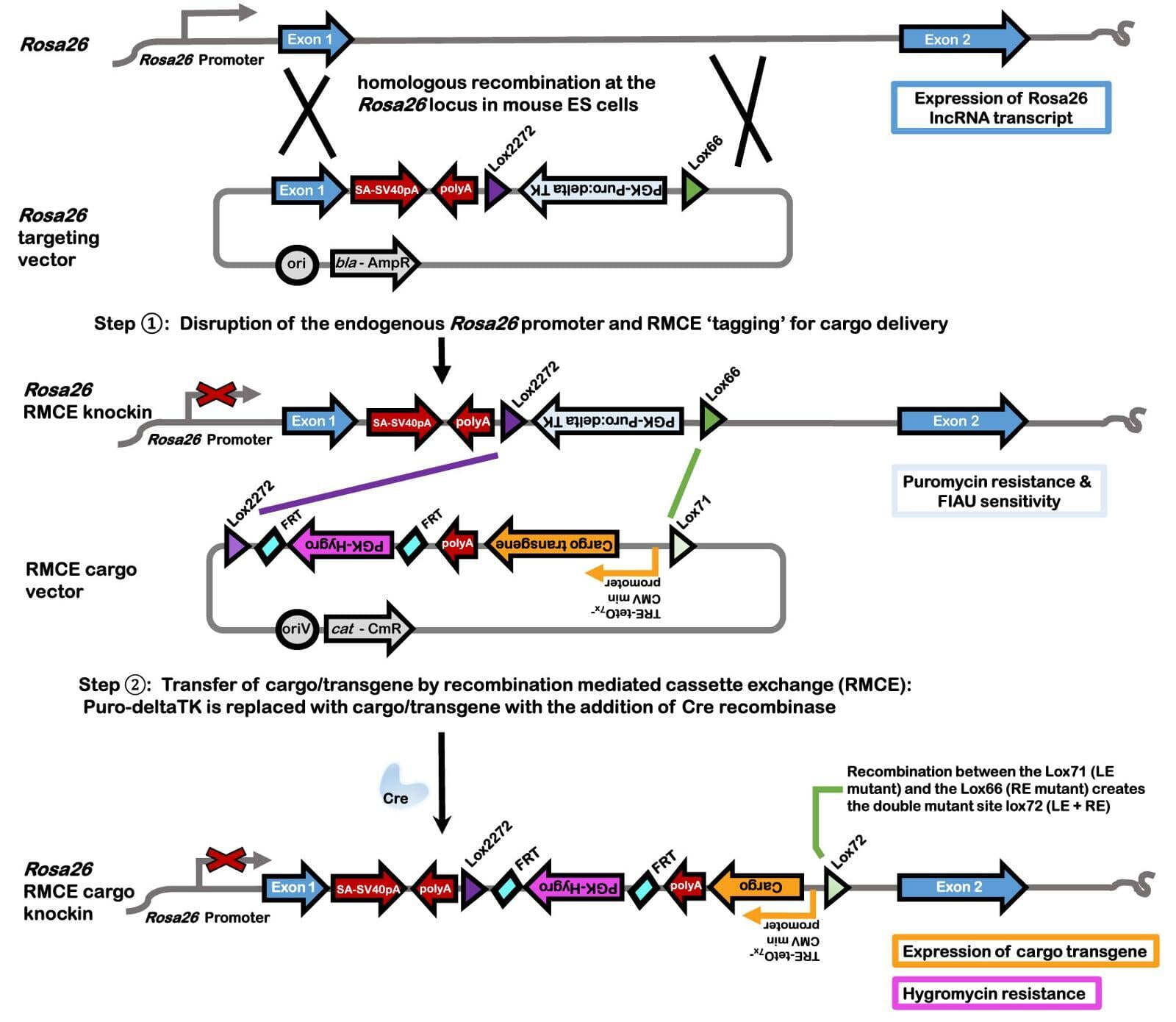
- Recombination mediated cassette exchange (RMCE) described in Lee et al, 2019⁶
In natural or synthetic contexts, gene drives often rely on some genetic element or circuit that can selfishly copy itself from one chromosome to another chromosome typically during early development or in germ cells. This selfish copying process known as an 'active genetic' allele spreads within a population at a higher frequency than traditional Mendelian inheritance. As such, this super-Mendelian inheritance is known as the gene drive. While gene drive systems have deployed homing nucleases such as I-SceI as described by Windbichler et al 2011⁷, more recently CRISPR-mediated gene drives have been shown as a proof-of-concept to work within the mouse female germline (Grunwald et al 2018⁸). Building on these milestones, the gene drive technology has the potential to transform the use of rodent biomedical models by reducing the number of matings needed to generate mutant offspring of desired alleles. Because the mutant allele can spread through a population rapidly, deployment of any gene drive requires a contemporaneous biocontainment strategy to mitigate spread to nontarget populations. One solution to the wild spreading of an allele is to deploy a local daisy-chain gene drive as described by Esvelt and Church (Noble et al 2019⁹). NovoHelix can assist the client to develop the gene drive and appropriate biocontainment strategy: please inquire with our scientific leadership team.