CRISPR tools
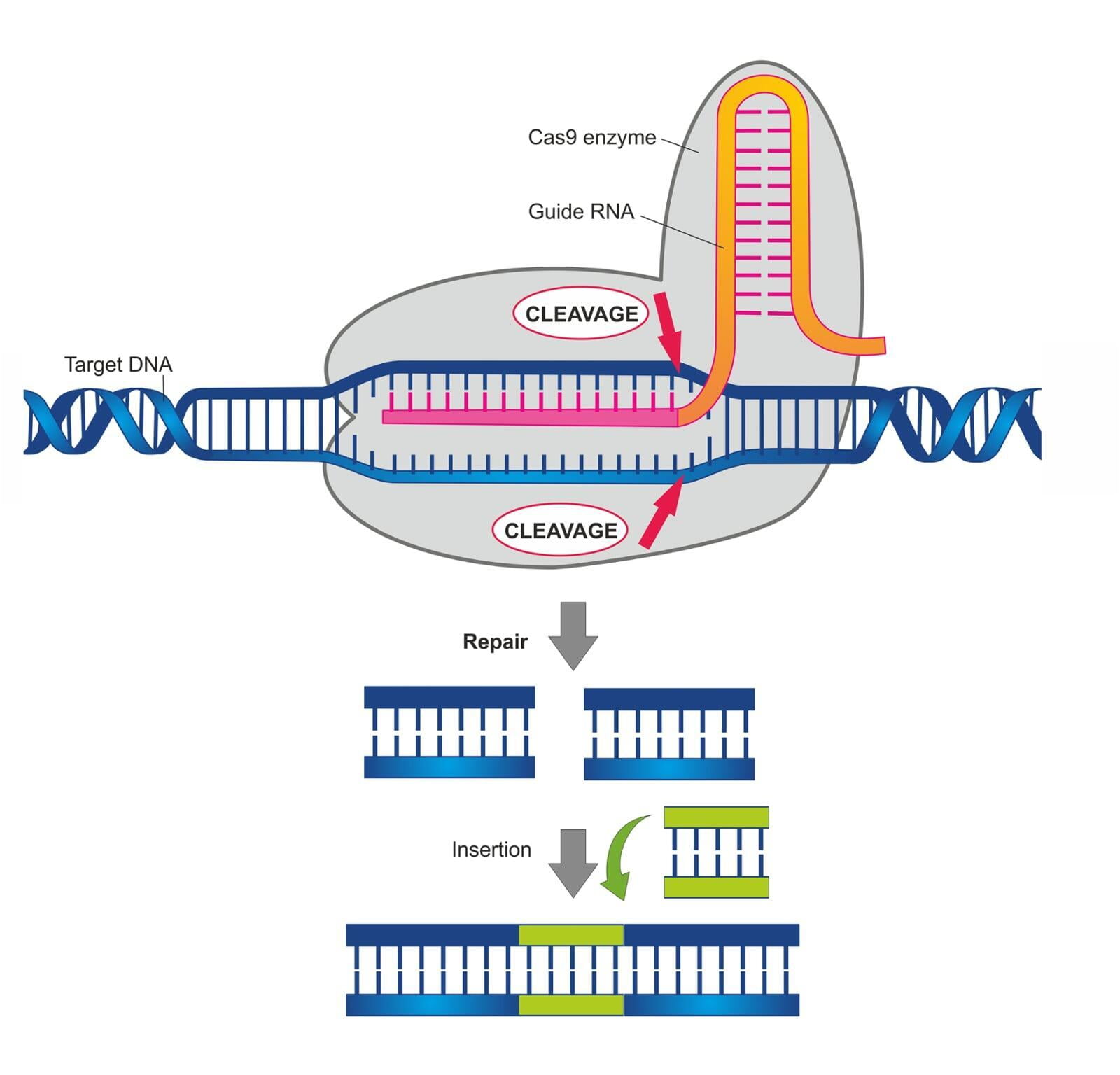
Prior to 2013, a handful of us were using protein engineering to build zinc finger nucleases/ZFN’s and subsequently TALEN’s (transcription activator-like effector nucleases) to modify mammalian genomes. The advent of gene editing from an RNA-guided programmable DNA cleavage system such as CRISPR-Cas has ushered in a new era of genetic engineering known as genome engineering (Mojica & Montoliu, Trends Microbiol. 2016¹). A typical gene editing experiment requires a target sequence and in some cases a repair template, a Cas endonuclease and RNA guide. The investigator must determine how these tools will be delivered to the model system and an appropriate biotype format such as plasmid DNA, RNA or ribonucleoprotein/RNP. For the generation of animal models by zygotic microinjection, we deploy Cas RNP complexes because they are immediately active upon delivery, and the RNPs have a limited half-life as compared to plasmids and thus reduce genetic mosaicism and reduce off-target cleavage events.
services
technology
references
forms
quote request
services
Service
Catalog Nr
Service Description
Price
Guide RNA design and cloning (plasmid DNA based)
MBCT001
1 target locus, up to 4 guide RNAs, U6 or T7 promoter vector
$1,775.00
Guide RNA design, cloning and validation (plasmid DNA based)
MBCT002
1 target locus, up to 4 guide RNAs, U6 or T7 promoter vector and validation by cell-based transfection.
$4,310.00
Development of guide RNAs - guide RNA cloning and design with T7 in vitro transcription (T7- IVT) to deliver in RNP format
MBCT003
Service is for design and in vitro transcription (IVT) of up to 6 guide RNAs for 1 target locust. Guides are delivered as ribonucleoprotein (RNP), consisting of the Cas protein in complex with a targeting gRNA.
$4,000.00
- Gene editing activity testing
- Format - cell-based transfection
- Assay - T7 endonuclease I/Cel-II/Surveyor
MBCT004
NovoHelix offers a gene editing service to help clients test their CRISPR tools including guide RNAs, high-performance mutant Cas proteins and base editors in plasmid DNA or RNP formats. A representative cell line will be transfected in triplicate and results will be generated by the mismatch-nucleases T7 Endo I or Cel-II as adopted from a protocol originally developed by Keith Joung's lab. Guide RNAs can also be tested in an vitro cutting assay if preferred, but we have found in vitro activity to supercede in vivo activity levels both in cellular and microinjection contexts; and hence in vitro mismatch nuclease assays often do not correlate to gene editing in vivo outcomes.
$4,750.00
- Gene editing activity testing
- Format - cell-based transfection
- Assay - genetic reporter & flow cytometry
MBCT005
NovoHelix offers a gene or base editing service to help clients test their CRISPR tools including guide RNAs, high-performance mutant Cas proteins and base editors in plasmid DNA or RNP formats. A representative cell line will be transfected in triplicate and results will be generated by a fluorescent reporter assay and flow cytometry.
$5,000.00
technology
references
Mojica FJM, Montoliu L. On the Origin of CRISPR-Cas Technology: From Prokaryotes to Mammals. Trends Microbiol. 2016 Oct;24(10):811-820. doi: 10.1016/j.tim.2016.06.005. Epub 2016 Jul 9. Review. PubMed PMID: 27401123.
Filippova J, Matveeva A, Zhuravlev E, Stepanov G. Guide RNA modification as a way to improve CRISPR/Cas9-based genome-editing systems. Biochimie. 2019 Dec;167:49-60. doi: 10.1016/j.biochi.2019.09.003. Epub 2019 Sep 4. Review. PubMed PMID: 31493470.
Palumbo CM, Gutierrez-Bujari JM, O'Geen H, Segal DJ, Beal PA. Versatile 3' Functionalization of CRISPR Single Guide RNA. Chembiochem. 2020 Jan 14. doi: 10.1002/cbic.201900736. [Epub ahead of print] PubMed PMID: 31943634.
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014 Mar;32(3):279-284. doi: 10.1038/nbt.2808. Epub 2014 Jan 26. PubMed PMID: 24463574; PubMed Central PMCID: PMC3988262.
Kocak DD, Josephs EA, Bhandarkar V, Adkar SS, Kwon JB, Gersbach CA. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat Biotechnol. 2019 Jun;37(6):657-666. doi: 10.1038/s41587-019-0095-1. Epub 2019 Apr 15. PubMed PMID: 30988504; PubMed Central PMCID: PMC6626619.
Kulcsár PI, Tálas A, Tóth E, Nyeste A, Ligeti Z, Welker Z, Welker E. Blackjack mutations improve the on-target activities of increased fidelity variants of SpCas9 with 5'G-extended sgRNAs. Nat Commun. 2020 Mar 6;11(1):1223. doi: 10.1038/s41467-020-15021-5. PubMed PMID: 32144253; PubMed Central PMCID: PMC7060260.
forms
quote request

