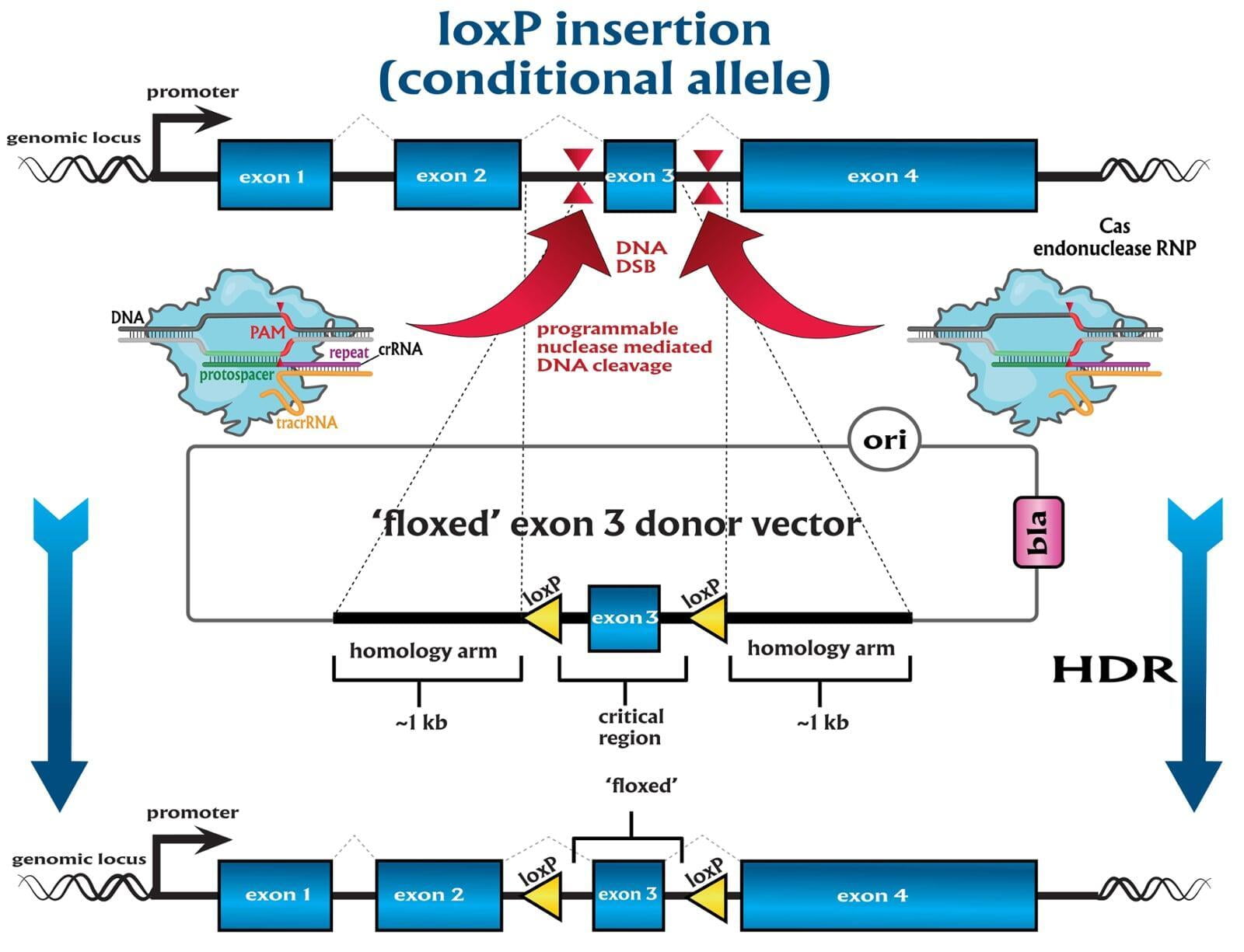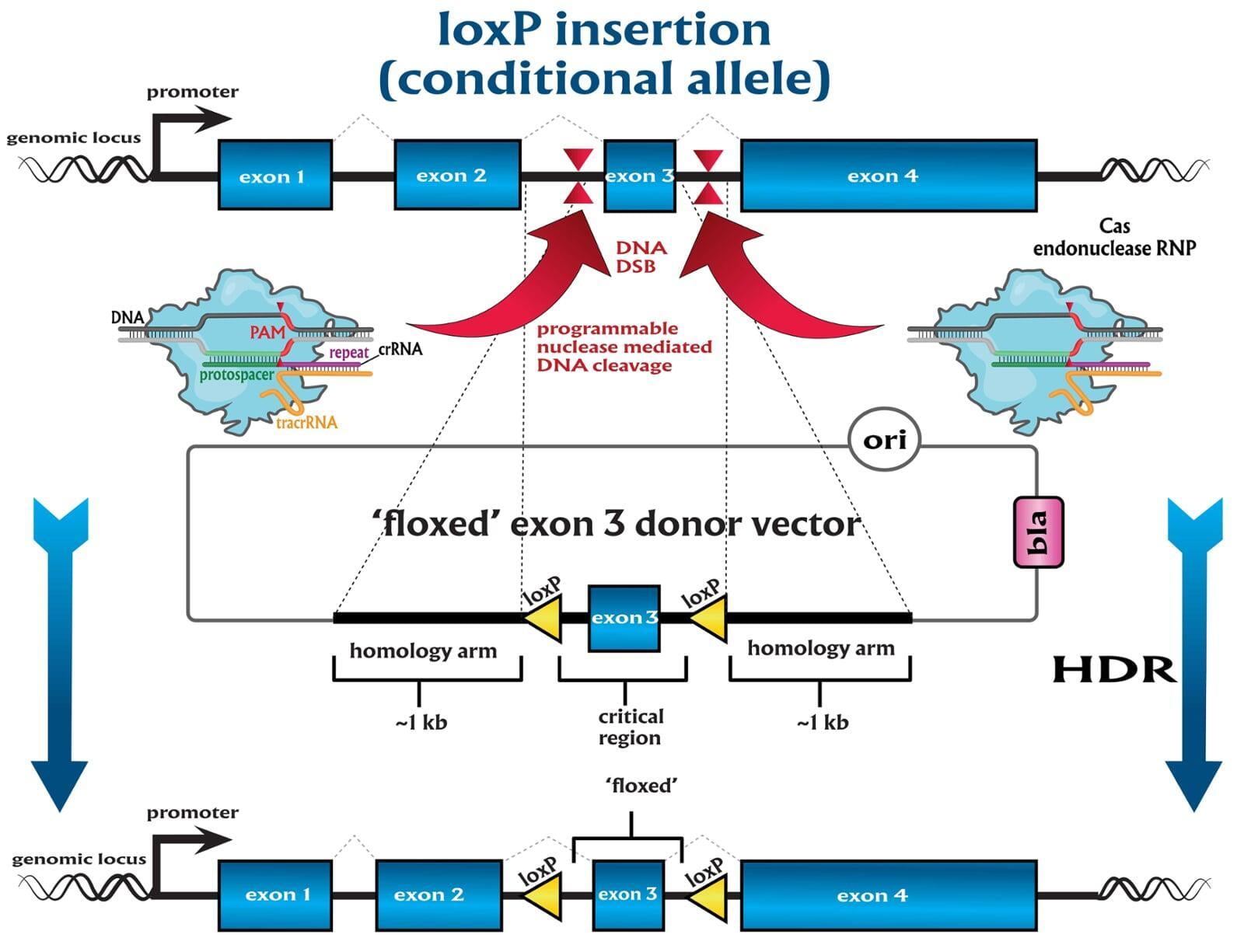
site-specific recombination technologies (SSR)
-conditional by inversion (COIN) as described by Economides et al, 2013¹
-flip-excision switches (FLEx) as described by Schnütgen et al, 2013²
-invertible intronic cassette (FLIP) as described by Andersson-Rolf et al, 2017³
-Recombination mediated cassette exchange (RMCE)
Service
Catalog Nr
Service Description
Timeline
Deliverables
Pricing
- Gene editing activity testing
- Format - cell-based transfection
- Assay - T7 endonuclease I/Cel-II/Surveyor
7-10 days
Gene editing activity of up to 6 guide RNAs
- Gene editing activity testing
- Format - cell-based transfection
- Assay - genetic reporter & flow cytometry
NovoHelix offers a gene editing service to help clients test their CRISPR tools including guide RNAs, high-performance mutant Cas proteins and base editors in plasmid DNA or RNP formats. A representative cell line will be transfected in triplicate and results will be generated by a fluorescent reporter assay and flow cytometry.
7-10 days
Gene editing activity of up to 6 guide RNAs
2-4 days
genotyping protocol
Service is for design and cloning of up to 6 guide RNAs for 1 target locust.
7-10 days
6 guide RNAs in plasmid vectors
7-10 days
6 guide RNAs ~ 500 ng/ul --- RNA format
Service
Catalog Nr
Service Description
Timeline
Deliverables
Pricing
- SSR allele
- C57BL/6 mouse via CRISPR microinjection
- guaranteed founders
The service includes donor vector or long single-stranded DNA (lssDNA) construction, guide RNA design using the latest design guidelines, i.e. full-length guides, chemically modified gRNAs, truncated sgRNAs, hp-sgRNAs (hairpin-sgRNAs), that produces high on-target activity in our surrogate cell-based gene editing assay while limiting off-target activity.
3 - 4 months founders; 5 - 8 months GLT F1s
At least 2 SSR founders (or germline transmitted F1s) with the confirmed mutation
- SSR allele
- C57BL/6 mouse via CRISPR microinjection
- non-guaranteed service (per injection session)
SSR knock-in mice production with microinjection of the repair template/Cas RNP complex performed as a per session service with a total of 500 zygotes injected. The service includes donor vector or long single-stranded DNA (lssDNA) construction, guide RNA design using the latest design guidelines, i.e. full-length guides, chemically modified gRNAs, truncated sgRNAs, hp-sgRNAs (hairpin-sgRNAs), that produces high on-target activity in our surrogate cell-based gene editing assay while limiting off-target activity.
3 - 4 months founders; 5 - 8 months GLT F1s
At least 500 embryos will be injected and implanted (2-3 microinjection sessions), which usually results in approximately 100 pups. Ear biopsies from all pups will be given to the investigator for genotyping. There is no guarantee that resulting pups will be gene edited and contain the SSR allele.
- SSR allele
- custom mouse strain - CRISPR-mediated
- guaranteed founders
SSR knock-in custom strain mice production via microinjection of the repair template/Cas RNP complex. The service includes donor vector or long single-stranded DNA (lssDNA) construction, guide RNA design using the latest design guidelines, i.e. full-length guides, chemically modified gRNAs, truncated sgRNAs, hp-sgRNAs (hairpin-sgRNAs), that produces high on-target activity in our surrogate cell-based gene editing assay while limiting off-target activity.
3 - 4 months founders; 5 - 8 months GLT F1s
At least 2 SSR founders (or germline transmitted F1s) with the confirmed mutation