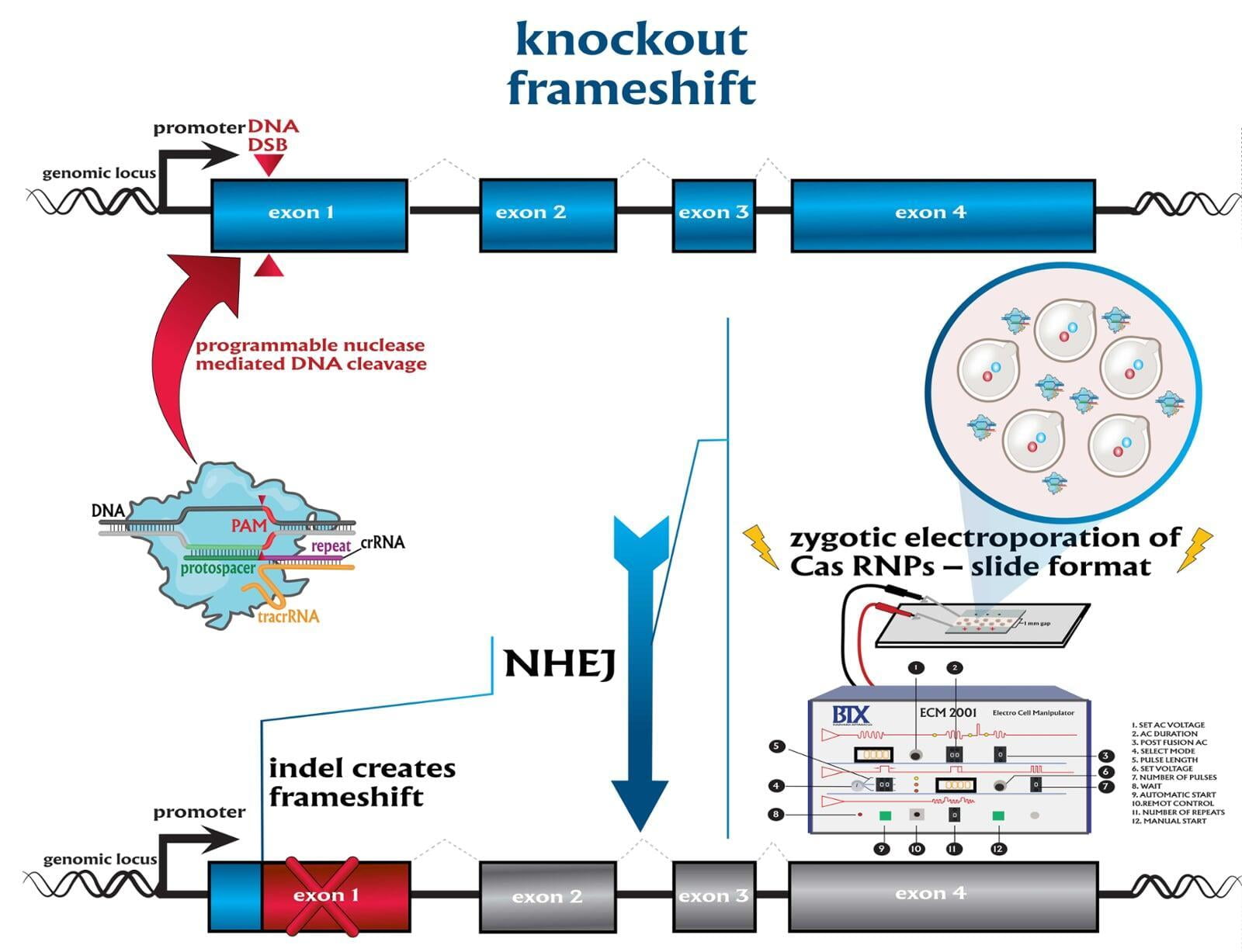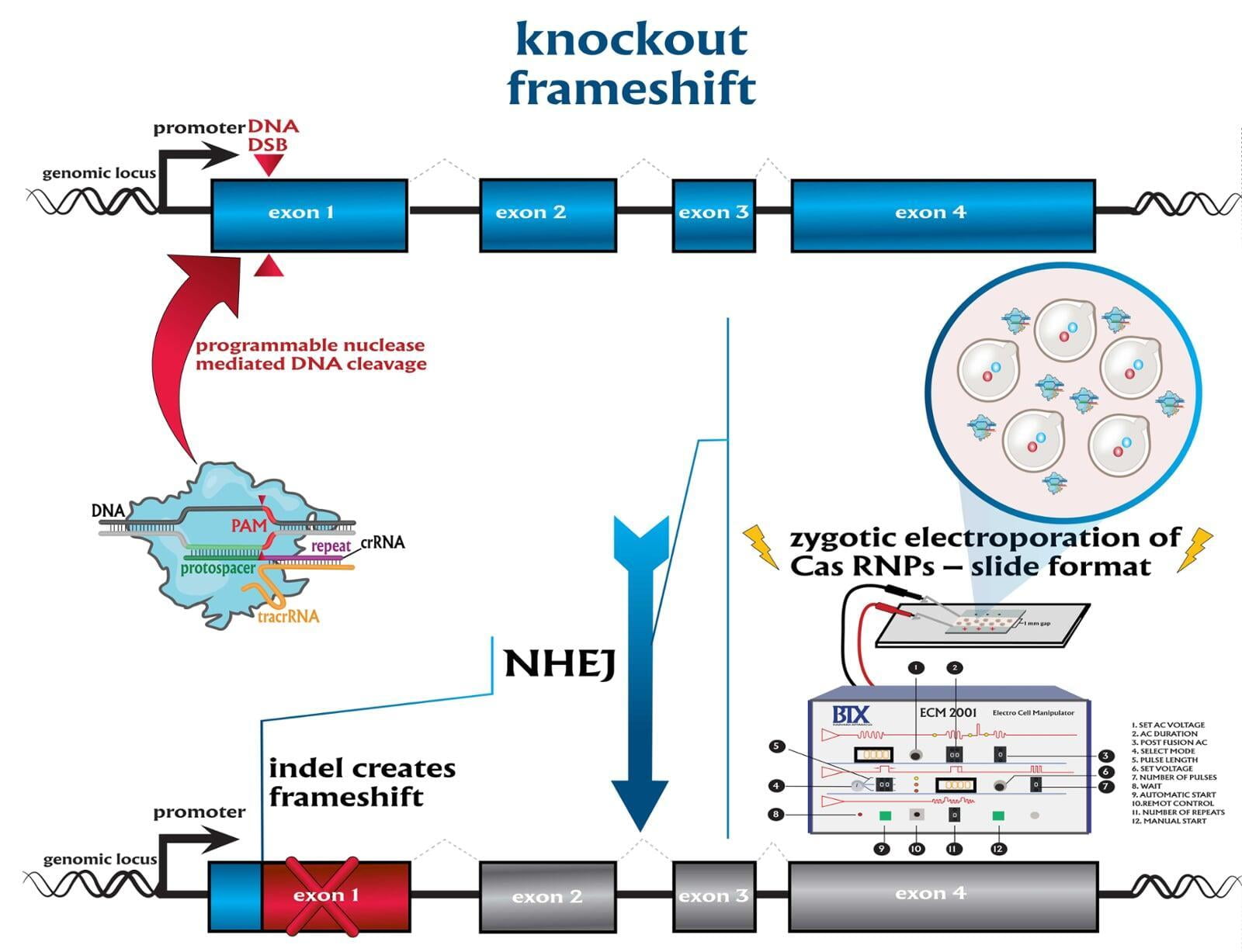
knockouts (KO)
To advance the study of gene function, NovoHelix offers a comprehensive loss-of-function gene platform to generate the desired animal model. CRISPR-nuclease assisted gene inactivation services include disruption of the translational reading frame such as frameshift stops (indels) to create simple knockouts, exonic deletions, single or multi-gene deletions, double knockouts (DKO), deletion of linked genes, megabase deletions and whole chromosomal ablation (induced aneuploidy). For knockout of multiple genes simultaneously also known as multiplexed genome engineering, the Cas12a (Cpf1) nuclease processes a single CRISPR array containing several target sites and disrupts the reading frames of multiple genes (Zetsche et al 2017, Campa et al 2019¹,² ). While this multiplexing technology is still in preliminary stages, NovoHelix has demonstrated expertise using cutting edge research tools to inactivate multiple genes and has entered the digital genome engineering era.
model generation
support services
technology
references
quote request
forms
model generation
Service
Catalog Nr
Service Description
Timeline
Deliverables
Pricing
- Simple frameshifting knockout
- C57BL/6 mouse via CRISPR microinjection
- guaranteed founders
KOMM001
The service includes disruption of the translational reading frame such as simple frameshifting mutations that cause a nonsense mutation, which results in a shorter protein chain because of an early stop codon. Design of a single effective guide RNA, TALEN or ZFN pair causes the protein's reading frame to be shifted via the introduction of a double-stranded DNA break and repair by non-homologous end joining (indels). The service includes guide RNA design using the latest design guidelines, i.e. full-length guides, chemically modified gRNAs, truncated sgRNAs, hp-sgRNAs (hairpin-sgRNAs), that produces high on-target activity in our surrogate cell-based gene editing assay while limiting off-target activity. For TALEN editing, we will build 3 obligate pairs and test activity for highest performing activity. For ZFNs, we will either build 3 obligate pairs or use a client-provided ZFN. We will consult with clients if TALEN or ZFN activity is insufficient by our surrogate gene activity assay after building 3 obligate heterodimers with the option to revert to a CRISPR-mediated strategy.
3 - 4 months founders; 5 - 8 months GLT F1s
At least 2 KO founders (or germline transmitted F1s) with the confirmed mutation
- Simple frameshifting knockout
- C57BL/6 mouse via CRISPR microinjection
- non-guaranteed service (per injection session)
KOMM002
The service includes disruption of the translational reading frame such as simple frameshifting mutations that cause a nonsense mutation, which results in a shorter protein chain because of an early stop codon. Design of a single effective guide RNA causes the protein's reading to be shifted via the introduction of a double-stranded DNA break and repair by non-homologous end joining (indels). Microinjection is performed as a per session service with a total of 500 zygotes injected.
3 - 4 months founders
At least 500 embryos will be injected and implanted (2-3 microinjection sessions), which usually results in approximately 100 pups. Ear biopsies from all pups will be given to the investigator for genotyping. There is no guarantee that resulting pups will be gene edited.
- Simple frameshifting knockout
- custom mouse strain - CRISPR-mediated
- guaranteed founders
KOMM003
The service includes disruption of the translational reading frame such as simple frameshifting mutations that cause a nonsense mutation, which results in a shorter protein chain because of an early stop codon. Design of a single effective guide RNA causes the protein's reading to be shifted via the introduction of a double-stranded DNA break and repair by non-homologous end joining (indels).
3 - 4 months founders; 5 - 8 months GLT F1s
At least 2 KO founders (or germline transmitted F1s) with the confirmed mutation
- Complex knockout (KO)
- C57BL/6 mouse via CRISPR microinjection
- guaranteed founders
KOMM004
Services included are disruption of the translational reading frame such as frameshift stops (indels), exon deletions, single or multi-gene deletions, double knockouts (DKO), deletion of linked genes, megabase deletions and whole chromosomal ablation (induced aneuploidy). At lease two founders are guaranteed.
3 - 4 months founders; 5 - 8 months GLT F1s
At least 2 KO founders (or germline transmitted F1s) with the confirmed mutation
- Complex knockout (KO)
- C57BL/6 mouse via CRISPR microinjection
- non-guaranteed service (per injection session)
KOMM005
Services included are disruption of the translational reading frame such as frameshift stops (indels), exon deletions, single or multi-gene deletions, double knockouts (DKO), deletion of linked genes, megabase deletions and whole chromosomal ablation (induced aneuploidy). For knockout of multiple genes simultaneously also known as multiplexed genome engineering, the Cas12a (Cpf1) nuclease processes a single CRISPR array containing several target sites and disrupts the reading frames of multiple genes. While this multiplexing technology is still in preliminary stages, NovoHelix has demonstrated expertise using custom nucleases to inactivate multiple genes.Microinjection is performed as a per session service with a total of 500 zygotes injected.
3 - 4 months founders
At least 500 embryos will be injected and implanted (2-3 microinjection sessions), which usually results in approximately 100 pups. Ear biopsies from all pups will be given to the investigator for genotyping. There is no guarantee that resulting pups will be gene edited.
- Complex knockout
- custom mouse strain - CRISPR-mediated knockin
- guaranteed founders
KOMM006
Services included are disruption of the translational reading frame such as frameshift stops (indels), exon deletions, single or multi-gene deletions, double knockouts (DKO), deletion of linked genes, megabase deletions and whole chromosomal ablation (induced aneuploidy). For knockout of multiple genes simultaneously also known as multiplexed genome engineering, the Cas12a (Cpf1) nuclease processes a single CRISPR array containing several target sites and disrupts the reading frames of multiple genes. While this multiplexing technology is still in preliminary stages, NovoHelix has demonstrated expertise using custom nucleases to inactivate multiple genes. At least two founders are guaranteed with this service.
3 - 4 months founders; 5 - 8 months GLT F1s
At least 2 KO founders (or germline transmitted F1s) with the confirmed mutation
support services
Service
Catalog Nr
Service Description
Timeline
Deliverables
Pricing
- Gene editing activity testing
- Format - cell-based transfection
- Assay - T7 endonuclease I/Cel-II/Surveyor
KOMM007
NovoHelix offers a gene editing service to help clients test their CRISPR tools including guide RNAs, high-performance mutant Cas proteins and base editors in plasmid DNA or RNP formats. A representative cell line will be transfected in triplicate and results will be generated by the mismatch-nucleases T7 Endo I or Cel-II as adopted from a protocol originally developed by Keith Joung's lab. Guide RNAs can also be tested in an vitro cutting assay if preferred, but we have found in vitro activity to supercede in vivo activity levels both in cellular and microinjection contexts; and hence in vitro mismatch nuclease assays often do not correlate to gene editing in vivo outcomes.
7-10 days
Activity of up to 6 guide RNAs
$3,000.00
- Gene editing activity testing
- Format - cell-based transfection
- Assay - genetic reporter & flow cytometry
KOMM008
NovoHelix offers a gene editing service to help clients test their CRISPR tools including guide RNAs, high-performance mutant Cas proteins and base editors in plasmid DNA or RNP formats. A representative cell line will be transfected in triplicate and results will be generated by a fluorescent reporter assay and flow cytometry.
7-10 days
Activity of up to 6 guide RNAs
$3,600.00
Genotyping Assay Development - frameshifting knockout
KOMM009
NovoHelix offers a knockout genotyping service to help clients develop robust genotyping protocols for screening animals after breeding and expansion of indidvidual mutant/KO founder lines.
2-4 days
genotyping protocol
$900.00
Genotyping Assay Development - complex knockouts
KOMM010
NovoHelix offers a knockout genotyping service to help clients develop robust genotyping protocols for screening animals after breeding and expansion of indidvidual mutant/KO founder lines.
5-7 days
genotyping protocol
$2,640.00
Development of guide RNAs - guide RNA cloning and design in a mammalian (U6, H1) expression vector or T7 in vitro transcription (T7- IVT) vector (DNA format)
KOMM011
Service is for design and cloning of up to 6 guide RNAs for 1 target locust in a DNA plasmid format.
7-10 days
6 guide RNAs in plasmid vectors
$1,800.00
Development of guide RNAs - guide RNA cloning and design with T7 in vitro transcription (T7- IVT) for use in RNP format (RNA format)
KOMM012
Service is for design and in vitro transcription (IVT) of up to 6 guide RNAs for 1 target locust.
7-10 days
6 guide RNAs ~ 500 ng/ul
$3,000.00
Client-provided guide RNAs - preparation for microinjection or slide electroporation (DNA/RNA format)
KOMM013
Service is for purification and for preparation of gRNAs for zygotic microinjection or slide-electroporation. Client should provide validated gRNA synthesis data such as small RNA electrophoresis results from Agilent's Bioanalyzer prior to microinjection and a minimum 200 ng/ul gRNA concentration. Purification is required to prevent the microinjection glass needle from clogging, to prevent embryo lysis and extensive delays in refitting and resetting microinjection setup.
$900.00
Client-provided dsDNA donor vector - preparation for microinjection
KOMM014
Purification of supercoiled plasmid by a proprietary anion exchange (AEX) chromatography, validation by restriction digestion and analysis by gel electrophoresis. Endotoxin levels are generally very low (0.05 – 0.5 ng LPS/µg) via our proprietary extraction method and are adequate for sensitive applications such as zygotic microinjection of mammalian embryos. The plasmid is predominantly in its supercoiled topology and free of RNA and protein contamination such as RNases and proteases. Drop dialysis in nucleofection/electroporation/microinjection buffer is included in the purification service. Validation by additional restriction digestions (greater than 3) is available with commensurate fee schedule. Purification is compulsory to prevent the microinjection glass needle from clogging, to prevent embryo lysis and extensive delays in refitting and resetting the microinjection setup.
$1,500.00
Mouse colony husbandry & management
KOMM015
mouse colony management including breeding, weaning, sampling, monitoring, per cage per day for the production of founders and F-generation pups
$3.60
technology
references
Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, van der Oost J, Zhang F. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017 Jan;35(1):31-34. doi: 10.1038/nbt.3737. Epub 2016 Dec 5. Erratum in: Nat Biotechnol. 2017 Feb 8;35(2):178. PubMed PMID: 27918548; PubMed Central PMCID: PMC5225075.
Campa CC, Weisbach NR, Santinha AJ, Incarnato D, Platt RJ. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat Methods. 2019 Sep;16(9):887-893. doi: 10.1038/s41592-019-0508-6. Epub 2019 Aug 12. PubMed PMID: 31406383.
Liu Z, Schiel JA, Maksimova E, Strezoska Ž, Zhao G, Anderson EM, Wu Y, Warren J, Bartels A, van Brabant Smith A, Lowe CE, Forbes KP. ErCas12a CRISPR-MAD7 for Model Generation in Human Cells, Mice, and Rats. CRISPR J. 2020 Apr;3(2):97-108. doi: 10.1089/crispr.2019.0068. PubMed PMID: 32315227.
quote request
forms