knock-in (KI) rat
Service
Catalog Nr
Service Description
Timeline
Deliverables
Pricing
- Custom knockin any locus
- Rat Sprague Dawley KI via CRISPR microinjection
- guaranteed founders
3 - 4 months founders; 5 - 8 months GLT F1s
At least 2 KI founders (or germline transmitted F1s) with the confirmed mutation
- Custom knockin any locus
- Rat Sprague Dawley KI via CRISPR microinjection
- non-guaranteed service (per injection session)
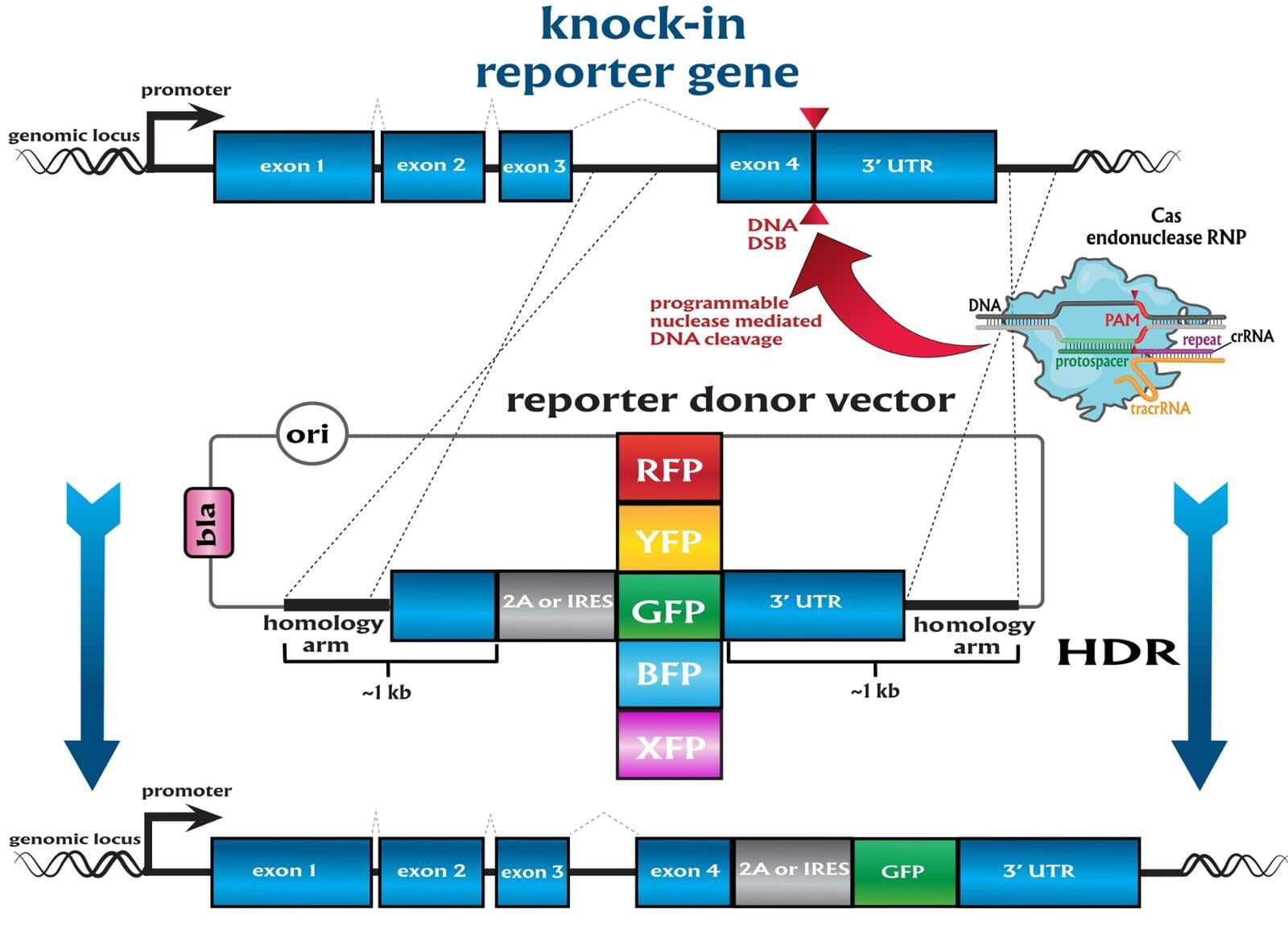
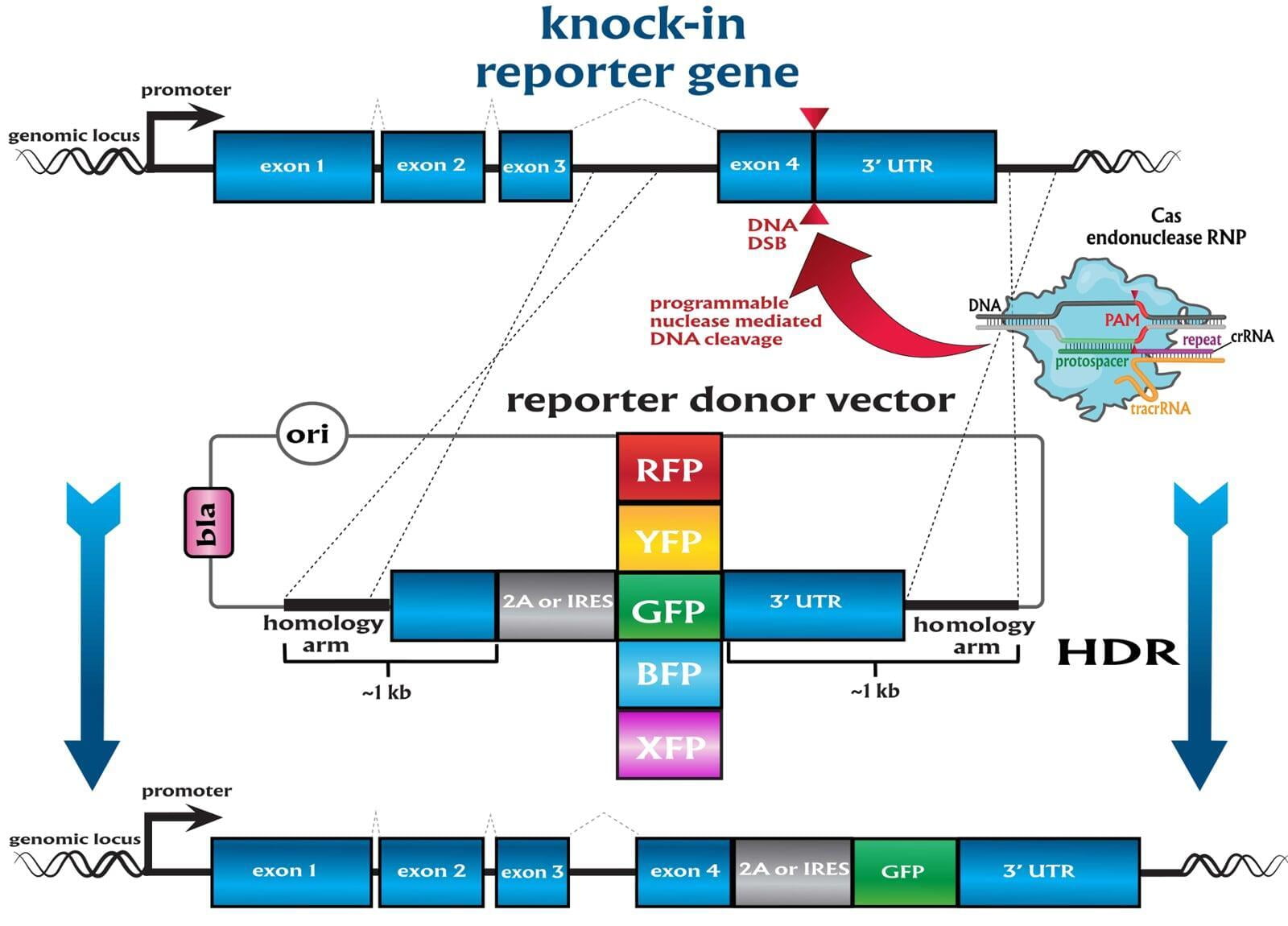
Services included are insertion of point mutations, altering amino acid codons, insertion of tags and barcodes, reporter gene knockins (GFP, mVenus, mCherry) and whole gene replacement such as humanization.
3 - 4 months founders
At least 500 embryos will be injected and implanted (2-3 microinjection sessions), which usually results in approximately 100 pups. Ear biopsies from all pups will be given to the investigator for genotyping. There is no guarantee that resulting pups will be gene edited.
- Custom knockin any locus
- custom rat strain - CRISPR-mediated knockin
- guaranteed founders
3 - 4 months founders; 5 - 8 months GLT F1s
At least 2 KI founders (or germline transmitted F1s) with the confirmed mutation
Service
Catalog Nr
Service Description
Timeline
Deliverables
Pricing
- Gene editing activity testing
- Format - cell-based transfection
- Assay - T7 endonuclease I/Cel-II/Surveyor
NovoHelix offers a gene editing service to help clients test their CRISPR tools including guide RNAs, high-performance mutant Cas proteins and base editors in plasmid DNA or RNP formats. A representative cell line will be transfected in triplicate and results will be generated by the mismatch-nucleases T7 Endo I or Cel-II as adopted from a protocol originally developed by Keith Joung's lab. Guide RNAs can also be tested in an vitro cutting assay if preferred, but we have found in vitro activity to supercede in vivo activity levels both in cellular and microinjection contexts; and hence in vitro mismatch nuclease assays often do not correlate to gene editing in vivo outcomes.
7-10 days
Activity of up to 6 guide RNAs
- Gene editing activity testing
- Format - cell-based transfection
- Assay - genetic reporter & flow cytometry
7-10 days
Activity of up to 6 guide RNAs
NovoHelix offers a knockout genotyping service to help clients develop robust genotyping protocols for screening animals after breeding and expansion of indidvidual mutant/KO founder lines.
2-4 days
genotyping protocol
5-7 days
genotyping protocol
Service is for design and cloning of up to 6 guide RNAs for 1 target locust in a DNA plasmid format.
7-10 days